We are thrilled to announce the release of WormBase ParaSite 18 (WBPS18), our most comprehensive update yet, with the largest number of new genomes and updated annotations since the resource was first launched! WBPS18 hosts 240 different genomes from 181 species.
Highlights
- Addition of 37 new genome assemblies for 14 existing and 12 new species, including a massive update of the trematode flukes genomes.
- Assembly and/or annotation updates for 3 genomes.
- Pairwise whole genome alignments are now available between all genomes in taxonomic groups.
- Integration of over 2 million AlphaFold 3D protein structures for 239 genomes.
- Updated RNA-Seq pipeline.
- Release 17 remains accessible.
- Mailing list.
New Species
- Aphelenchoides besseyi is a plant-parasitic nematode that causes root-knot disease in a variety of crops.
- Aphelenchoides bicaudatus is a plant-parasitic nematode that causes damage to a variety of crops, including potatoes, tomatoes, and strawberries.
- Aphelenchoides fujianensis is a plant-parasitic nematode that causes damage to a variety of crops, including rice, wheat, and soybeans.
- Caenorhabditis auriculariae is a free-living nematode that is used as a model organism in scientific research.
- Meloidogyne chitwoodi is a plant-parasitic nematode that causes root-knot disease in a variety of crops.
New Trematodes (Flukes)
- Dicrocoelium dendriticum is a trematode fluke that is a parasite of ruminants, including cattle, sheep, and goats.
- Heterobilharzia americana is a trematode fluke that is a parasite of humans and other mammals.
- Schistosoma guineensis is a trematode fluke that is a parasite of humans and other primates.
- Schistosoma intercalatum is a trematode fluke that is a parasite of humans and other primates.
- Schistosoma spindale is a trematode fluke that is a parasite of humans and other primates.
- Schistosoma turkestanicum is a trematode fluke that is a parasite of humans and other mammals.
- Trichobilharzia szidati is a trematode fluke that is a parasite of humans and other mammals.
Genome Updates
- Caenorhabditis briggsae: 2 new long-read sequenced assemblies with RNASeq-assisted (long and short read sequencing data) annotations were imported for QX1410 and VX34.
- Dirofilaria immitis: A new long-read sequenced assembly.
- Globodera pallida: 2 new long-read sequenced assemblies with RNASeq-assisted annotations were imported for Newton and D383 strains.
- Heterodera schachtii: A new long-read sequenced assembly with RNASeq-assisted gene annotation.
- Parapristionchus giblindavisi: A new long-read sequenced assembly with RNASeq-assisted gene annotation also based on the community-curated gene annotations of P. pacificus.
- Pristionchus exspectatus: A new long-read sequenced assembly with RNASeq-assisted gene annotation also based on the community-curated gene annotations of P. pacificus.
- Schmidtea mediterranea: A new phased long-read sequenced assembly with RNASeq-assisted annotation. Both haplotypes are available: Haplotype 1, Haplotype 2. Also, low confidence gene models from the same annotation source have been added as Jbrowse tracks (Haplotype 1 Jbrowse, Haplotype 2 Jbrowse) and can be loaded from the left menu. You can use the Jbrowse search bar to search for these genes.
- Angiostrongylus costaricensis: A new alternative set of gene models from da Silva EMG et al., 2022, for the Crissiumal strain has been added as Jbrowse tracks (Jbrowse) and can be loaded from the left menu. You can use the Jbrowse search bar to search for these genes.
Trematodes (Flukes)
- Schistosoma bovis: Two new long-read sequenced assemblies with RNASeq-assisted annotation: tdSchBovi1.1 and tdSchBovi2.1.
- Schistosoma curassoni: A new long-read sequenced assembly with RNASeq-assisted annotation.
- Schistosoma haematobium (UoM_Shae.V3): An update to the previous SchHae_2.0 reference genome. The new reference is long-read sequenced with RNASeq-assisted gene annotation.
- Schistosoma haematobium (PRJEB44434): Two new long-read sequenced assemblies with RNASeq-assisted annotation: tdSchHaem1.1 and tdSchHaem2.1.
- Schistosoma mansoni: Following incorporation of HiC data, and further PacBio analysis to resolve repeats a near-complete chromosomal assembly v10 is now available. Gene models were generated using RNASeq (including IsoSeq) data. More than 4500 genes have been manually curated.
- Schistosoma margrebowiei: A new long-read sequenced assembly with RNASeq-assisted (including IsoSeq) annotation.
- Schistosoma mattheei: A new long-read sequenced assembly with RNASeq-assisted (including IsoSeq) annotation.
- Schistosoma rodhaini: Two new long-read sequenced assemblies with RNASeq-assisted (including IsoSeq) annotation: tdSchRodh1.1 and tdSchRodh2.1.
- Trichobilharzia regenti: A new long-read sequenced assembly with RNASeq-assisted (including IsoSeq) annotation.
Pairwise Whole Genome Alignments
In WBPS18, users can now browse and visualise whole genome alignments between genomes in the same taxonomic group. Cactus, a next-generation aligner that stores whole-genome alignments in a graph structure, was utilised to calculate multiple pairwise genome alignments between WBPS genomes. These genomes have been assigned to 5 big taxonomic groups, and pairwise alignments can be viewed between the genomes of each group.
| Taxonomic Group Identifier | Genomes |
|---|---|
| Anc001 | Platyhelminthes |
| Anc005 | Clade I nematodes |
| Anc020 | Clade III nematodes |
| Anc028 | Clade V nematodes |
| Anc029 | Clade IV nematodes |
These alignments are viewable through the Ensembl Genome Browser. Follow this tutorial to learn how to browse these alignments.
Fetching genome alignments is a resource-consuming process, so please be patient while you are trying to load the alignment tracks and hit the refresh buttons in case any errors appear.
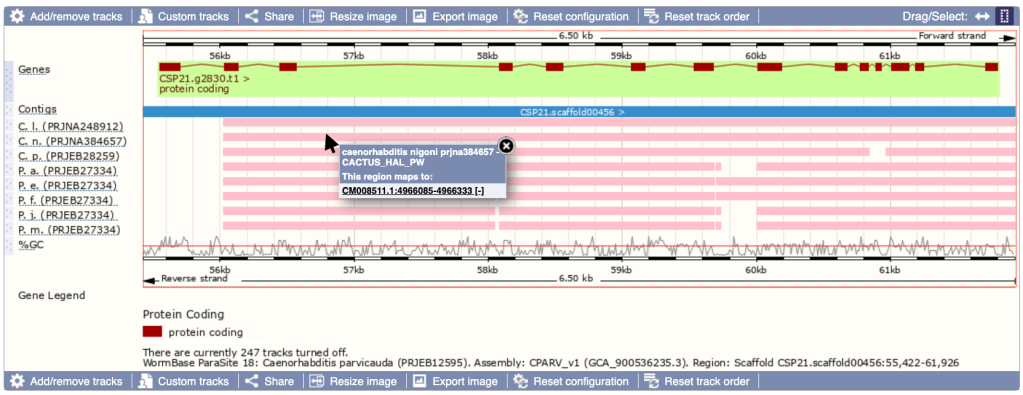
Ensembl Genome Browser view with pairwise alignment tracks in WBPS18. The view focuses on the genomic region of the Caenorhabditis parvicauda CSP21.g2830 gene. Aligned genomic regions of other Caenorhabditis and Pristionchus genomes have been loaded (pink tracks).
The whole genome alignments are also available for download in HAL file format, through our FTP site.
Full integration of AlphaFold models
In July 2022, AlphaFold predicted structures for almost every catalogued protein known to science.
In release 18, WormBase ParaSite incorporated these models. Now, over 2 million 3D protein structure models from AlphaFold are available for 239 genomes species. The AlphaFold predicted model (example) is available through the top-left menu on the transcript pages.
Follow this tutorial to learn how to browse your favourite protein models in WormBase ParaSite.
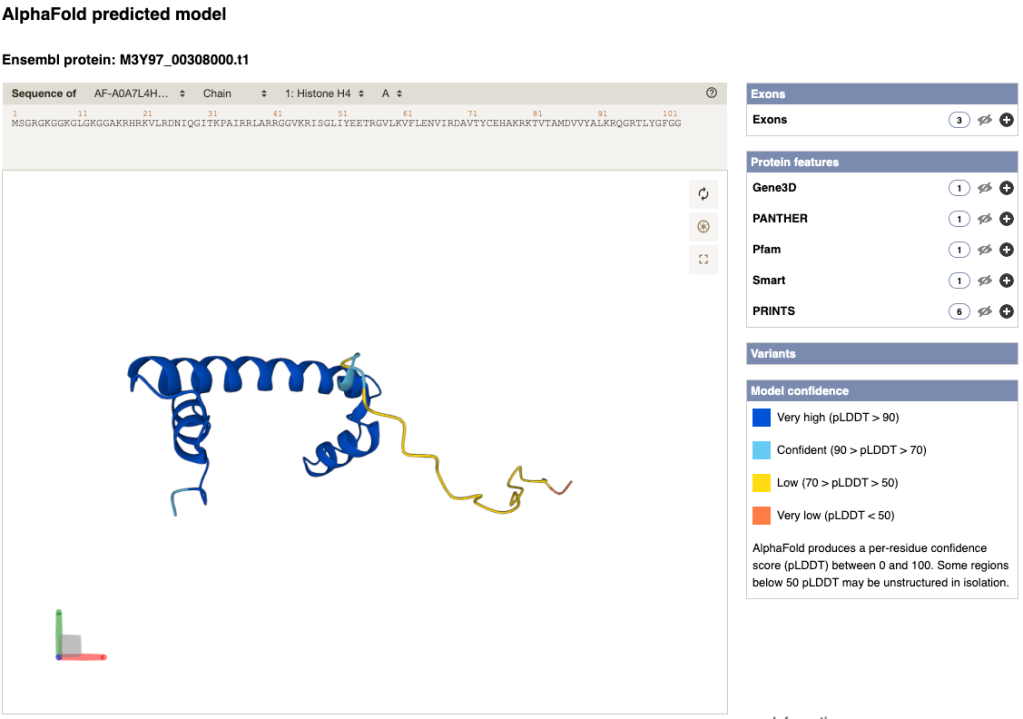
AlphaFold widget in WBPS18. The protein shown here corresponds to the M3Y97_00308000 gene of the newly added Aphelenchoides bicaudatus genome.
Updated RNA-Seq data
From release 16, due to the retirement of the RNASeq-er API, RNASeq data of genomes that had assembly or annotation updates were not re-aligned. For 34 updated genomes (releases 16 and 17), this affected their RNA-Seq alignment tracks available on the Ensembl Browser, Jbrowse and their gene counts on the Gene expression platform.
For this release, we utilised the novel RNA-Seq pipeline developed by the Ensembl Metazoa group to re-align the RNA-Seq data for these 34 genomes. The pipeline is using HISAT2 to perform the read alignments and htseq-count to count reads in features.
If you would like a new gene expression study to be included, please contact us.
“How can I browse RNA-Seq data in WormBase ParaSite?”
- Browse our gene expression pages (use the “Gene Expression” button in the “Navigation” box on the genome landing page of each genome) to explore the gene expression of your gene of interest across different samples and perform differential expression analysis.
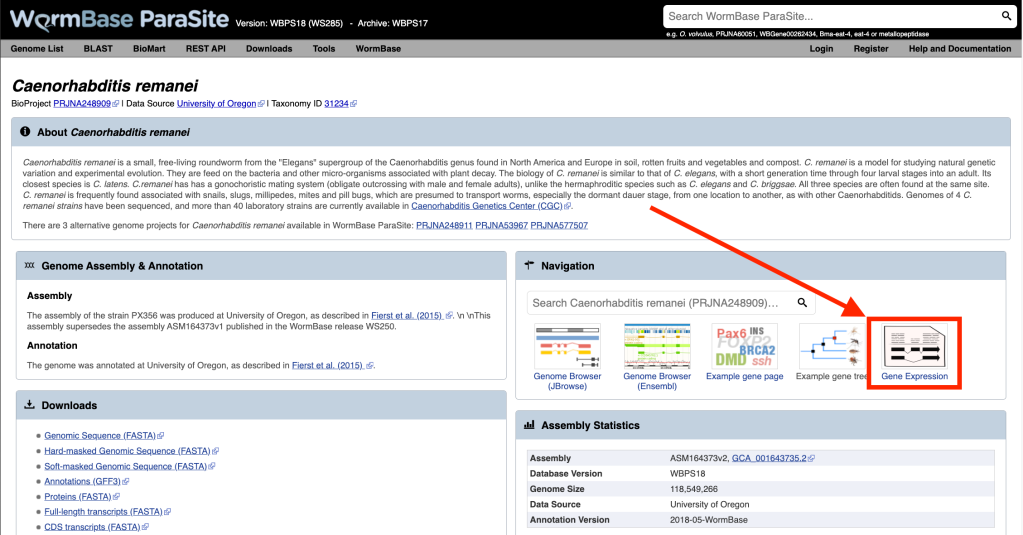
- Use Jbrowse genome browser, to visualise RNA-Seq samples as genome browser tracks.
- Use Genome Browser (Ensembl), to visualise RNA-Seq samples as genome browser tracks.
Archiving Site
Our previous Release 17 has been archived and remains accessible here.
Subscribe to our new mailing list
If you’re using WormBase ParaSite, then you should subscribe to our mailing list so you can frequently receive updates on WormBase Parasite new releases, like this one, new features, career opportunities, and upcoming conferences and workshops.
Subscribing to the WormBase Parasite mailing list is easy! Simply fill out the subscription form at https://forms.gle/gN5NqZJ3FeVDkpTA8 and stay updated with our latest news.
Problems/issues
If you encounter any problems with latest release, including broken links or missing genes, please don’t hesitate to contact us.